Hydroxyazole scaffold-based Plasmodium falciparum dihydroorotate dehydrogenase inhibitors: Synthesis, biological evaluation and X-ray structural studies.
Pippione, A.C., Sainas, S., Goyal, P., Fritzson, I., Cassiano, G.C., Giraudo, A., Giorgis, M., Tavella, T.A., Bagnati, R., Rolando, B., Caing-Carlsson, R., Costa, F.T.M., Andrade, C.H., Al-Karadaghi, S., Boschi, D., Friemann, R., Lolli, M.L.(2018) Eur J Med Chem 163: 266-280
- PubMed: 30529545
- DOI: https://doi.org/10.1016/j.ejmech.2018.11.044
- Primary Citation of Related Structures:
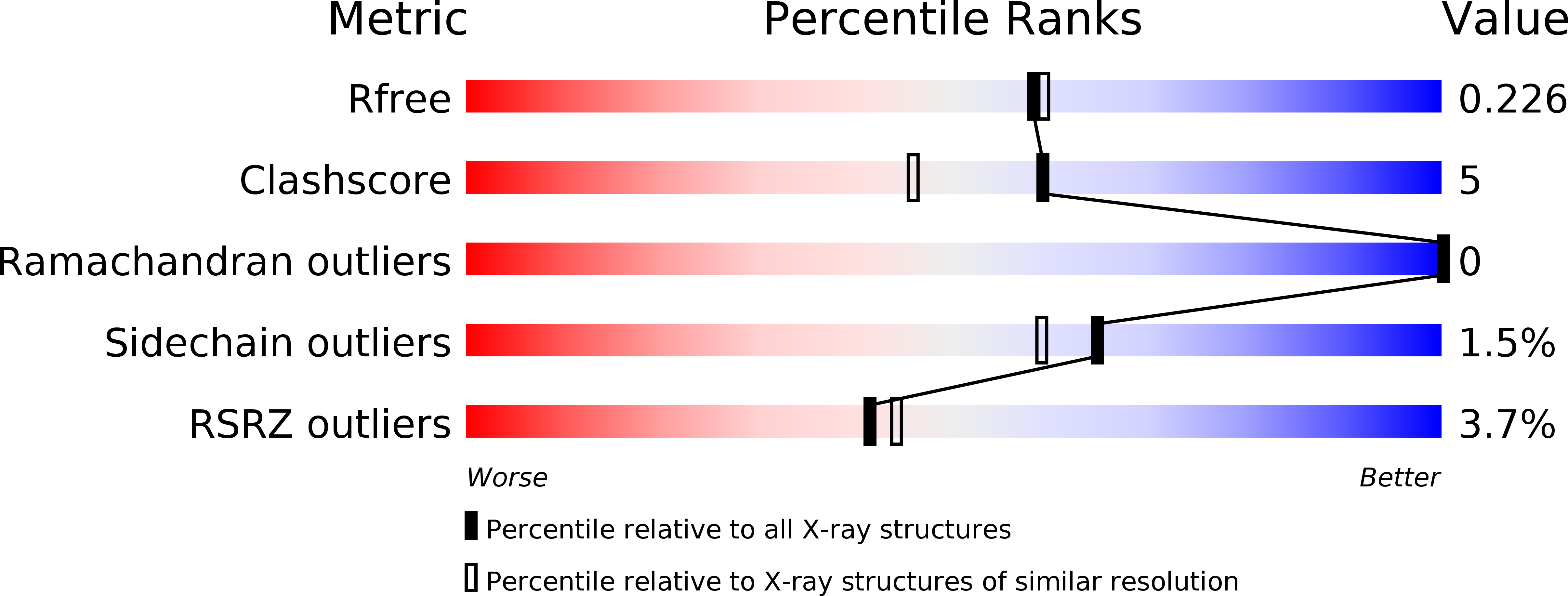
6I4B, 6I55 - PubMed Abstract:
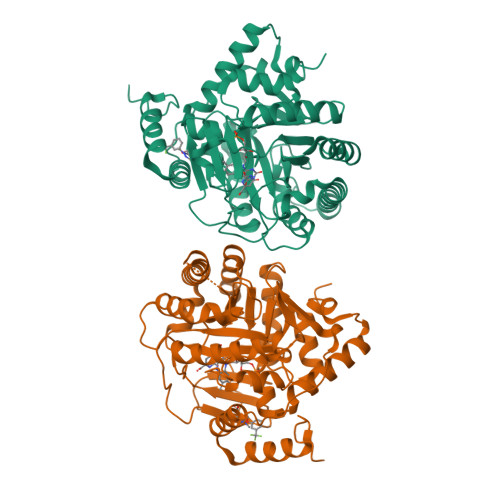
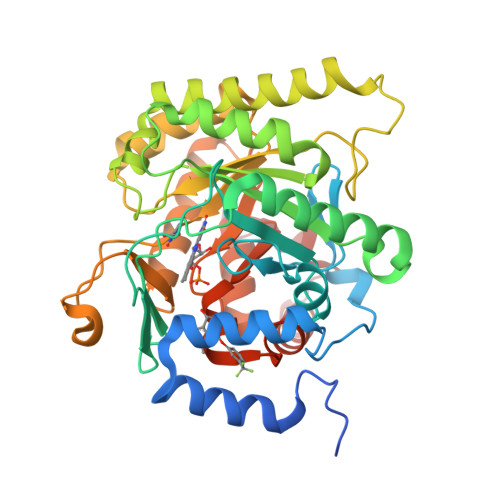
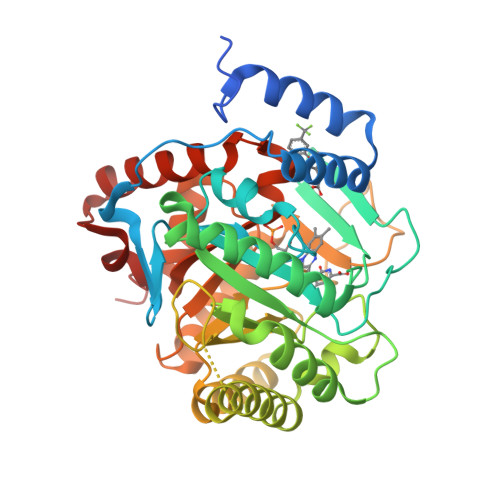
Plasmodium falciparum dihydroorotate dehydrogenase (PfDHODH) has been clinically validated as a target for antimalarial drug discovery, as a triazolopyrimidine class inhibitor (DSM265) is currently undergoing clinical development. Here, we have identified new hydroxyazole scaffold-based PfDHODH inhibitors belonging to two different chemical series. The first series was designed by a scaffold hopping strategy that exploits the use of hydroxylated azoles. Within this series, the hydroxythiadiazole 3 was identified as the best selective PfDHODH inhibitor (IC 50 12.0 μM). The second series was designed by modulating four different positions of the hydroxypyrazole scaffold. In particular, hydroxypyrazoles 7e and 7f were shown to be active in the low μM range (IC 50 2.8 and 5.3 μM, respectively). All three compounds, 3, 7e and 7f showed clear selectivity over human DHODH (IC 50 > 200 μM), low cytotoxicity, and retained micromolar activity in P. falciparum-infected erythrocytes. The crystallographic structures of PfDHODH in complex with compounds 3 and 7e proved their binding mode, supplying essential data for future optimization of these scaffolds.
Organizational Affiliation:
Department of Science and Drug Technology, University of Turin, via Pietro Giuria 9, 10125, Turin, Italy.